
On April 17, Si Tong's research group of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published a research paper in ACS Synthetic Biology, an international academic journal, under the title of “Deep Mutational Scanning of an Oxygen-Independent Fluorescent Protein CreiLOV for Comprehensive Profiling of Mutational and Epistatic Effects”. The non-oxygen dependent fluorescent protein CreiLOV is an important tool for studying anaerobic biological systems. The research team relied on the major technological infrastructure of synthetic biology research in Shenzhen, characterized the sequence - function relationship based on the FACS-seq method, and constructed a combinatorial saturation mutagenesis library containing 20 mutations at 15 loci (The capacity of the theoretical library is 184,000). By using a machine learning model, the research team predicted the high-order combinatorial mutagenesis effects between 15 loci based on a small amount of low-order (two - two, three - three) mutagenesis data; in the optimal case, only 0.25% of the experimental data of the theoretical design space were required to be covered to achieve reliable prediction of all combinatorial mutagenesis spaces. This study provides novel tools, empirical data, and theoretical guidance for machine learning-aided protein engineering practices. Dr. Chen Yongcan, an assistant researcher of the research group, is the first author of the article, and researcher Si Tong and assistant researcher Zhang Jianzhi are the corresponding authors of the article. This paper was included in AI for Synthetic Biology, a Virtual Special Issue organized by Professor Huimin Zhao, the editor in chief of the journal.

Screenshot of the published paper
Link of the paper: https://pubs.acs.org/doi/10.1021/acssynbio.2c00662
The maturation process of the chromophores of the conventional green fluorescent protein (GFP) relies on oxygen and cannot be applied to the study of biological systems such as gut microbiota, tumor interior, and anaerobic fermentation. Unlike GFP, the maturation of the chromophores of Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) does not require oxygen and has significant potential in studying anaerobic biological processes. FbFP originates from the light-oxygen-voltage domain (LOV domain) of photosensitive proteins. When the natural LOV domain is excited by blue or ultraviolet light, FMN and a conserved cysteine residue in the binding pocket of this domain form a covalent adduct, accompanied by fluorescence disappearance and conformational changes; in dark environment, the covalent adduct decays, and fluorescence recovers. When the cysteine mutates to alanine, the LOV domain can be transformed into a stable FbFP with a maximum fluorescence emission wavelength of 495 nm. FbFP has the advantages of low molecular weight, monosomy, fast maturation of chromophores, high pH and high thermal stability. However, the fluorescence intensity and quantum yield of FbFP are lower than those of GFP, so protein engineering is required. Previously, conventional directed evolution methods such as error-prone PCR and site-specific mutagenesis were usually used for engineering modifications of FbFP, by which the sequence space was less explored.
Deep mutational scanning (DMS) can be used to systematically analyze the sequence - function relationship of protein mutants by integrating large-scale mutagenesis library construction, high-throughput screening, and NGS sequencing. At present, a large number of protein engineering studies use single locus saturation mutational scanning libraries for deep mutational scanning, which greatly expands the mutation loci and types. However, good protein performance often requires the introduction of a number of amino acid mutations. There may be epistasis between mutations, where the effects of two or more mutations are different from the sum of their respective effects. Therefore, even if all single locus mutagenic effects are known, rational design of multi-loci mutations is challenging. Although dominant mutation combinations can accumulate through multiple rounds of directed evolution, this greedy strategy may fall into local optimum due to possible epistasis of sign or bidirectional sign between mutations.
In this study, the author used CreiLOV from Chlamydomonas reinhardtii as the research object and constructed a single locus saturation mutagenesis library with 118 loci by using NNK degenerate codons (The theoretical library capacity is 2,360). In order to obtain sequence - fluorescence intensity data, fluorescence activated cell sorting sequencing (FACS-seq) and phenotype estimation methods were used for rapid characterization. The filtered 2,185 mutant sequences accounted for over 92% of the theoretical library capacity. The author also compared a number of phenotypic estimation methods, and the results showed that each biological duplication correlation and estimation and measurement correlation obtained by using the simple weighted averages method was the highest, followed by the maximum likelihood estimation based on Gamma distribution and normal distribution. Based on sequence - fluorescence intensity data, the author identified key loci, regions, and amino acid mutations that attenuated or enhanced the fluorescence intensity of CreiLOV (Figure 1).
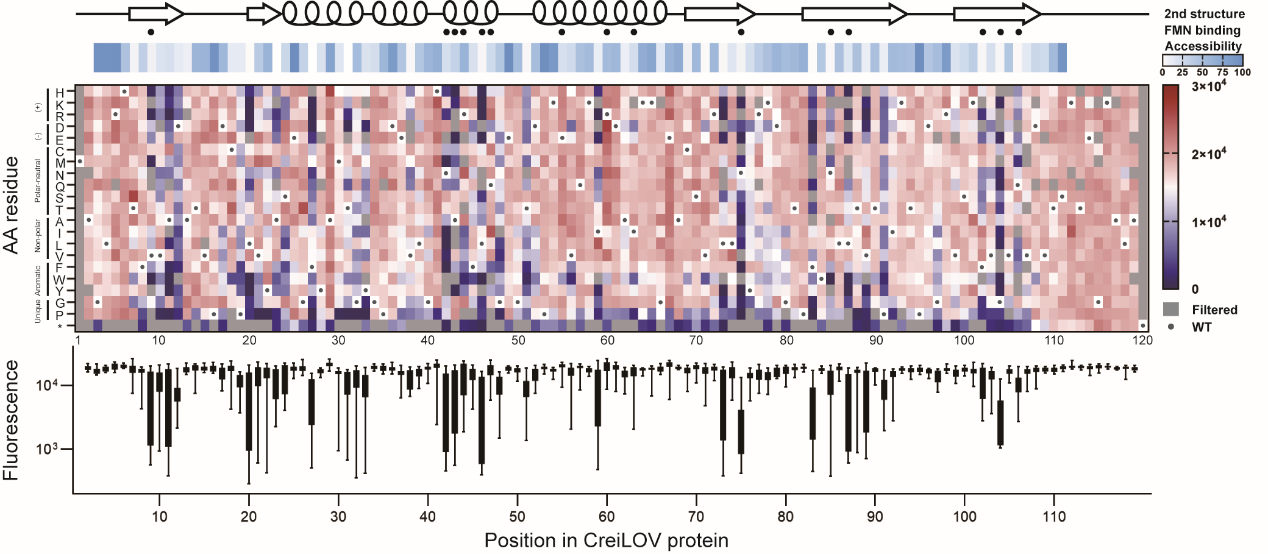
Figure 1 Analysis of CreiLOV single locus mutagenic effect
Based on the results of single locus saturation mutational scanning, the author further constructed a saturation mutation combination library covering 20 amino acid mutations at 15 loci with a theoretical library capacity of 184,000. Using a larger scale FACS-seq, the author analyzed the sequence - fluorescence intensity relationship of multi-loci mutants, and the filtered 165,000 mutant sequences accounted for about 90% of the theoretical library capacity. The author found that the overall fluorescence intensity gradually attenuated with the increase of the number of mutation loci. Since all mutations had fluorescence enhancement or neutral effects, it indicated there was extensive negative epistasis. The statistical analysis of specific epistasis between amino acid mutations demonstrated this inference (Figure 2).
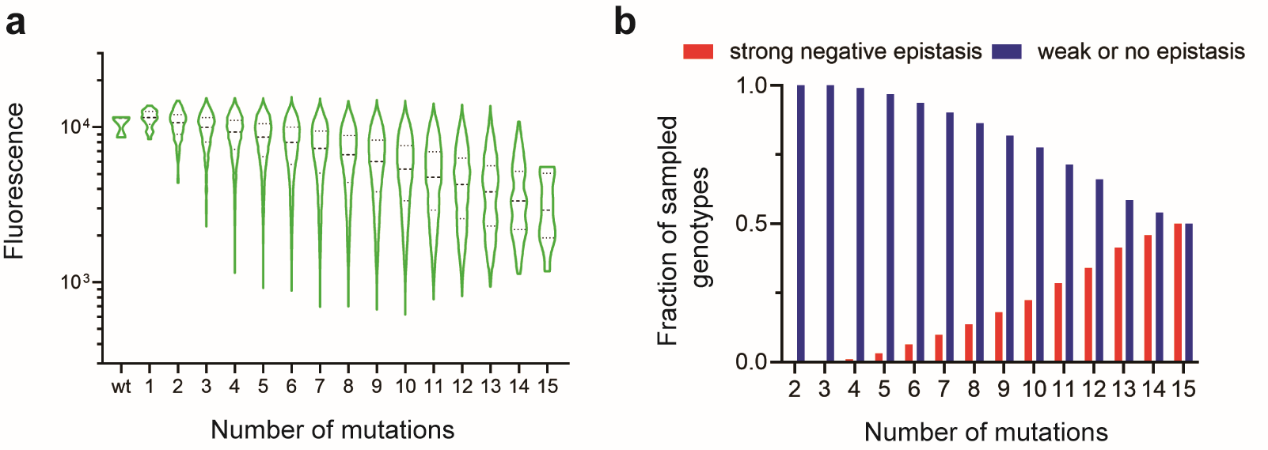
Figure 2 (a) Phenotypic distribution of combinatorial mutant and (b) Analysis of specific epistasis of CreiLOV
In recent years, scientists have found that the interpretation of mutagenic effect is also influenced by nonspecific epistasis, also known as global epistasis. Nonspecific epistasis is a universal feature of genotype-phenotype map (G-P map) due to the nonlinear relationship between physical properties and biological effects. Ignoring this nonlinear relationship often leads to overestimation of specific epistasis. MAVE-NN is a recently developed quantitative modeling strategy that integrates genotype-phenotype map model, global epistasis model, and noise model, and measures model performance using three mutual information indexes from information theory. The author used MAVE-NN to quantitatively model the combinatorial saturation mutagenesis dataset, and found that both the additive model based on genotype-phenotype map and the black box model showed an S-shaped relationship between the potential phenotype of CreiLOV and the measurements. Given this nonlinear feature, the predicted values of the model are highly correlated with the experimental measurements (Figure 3).
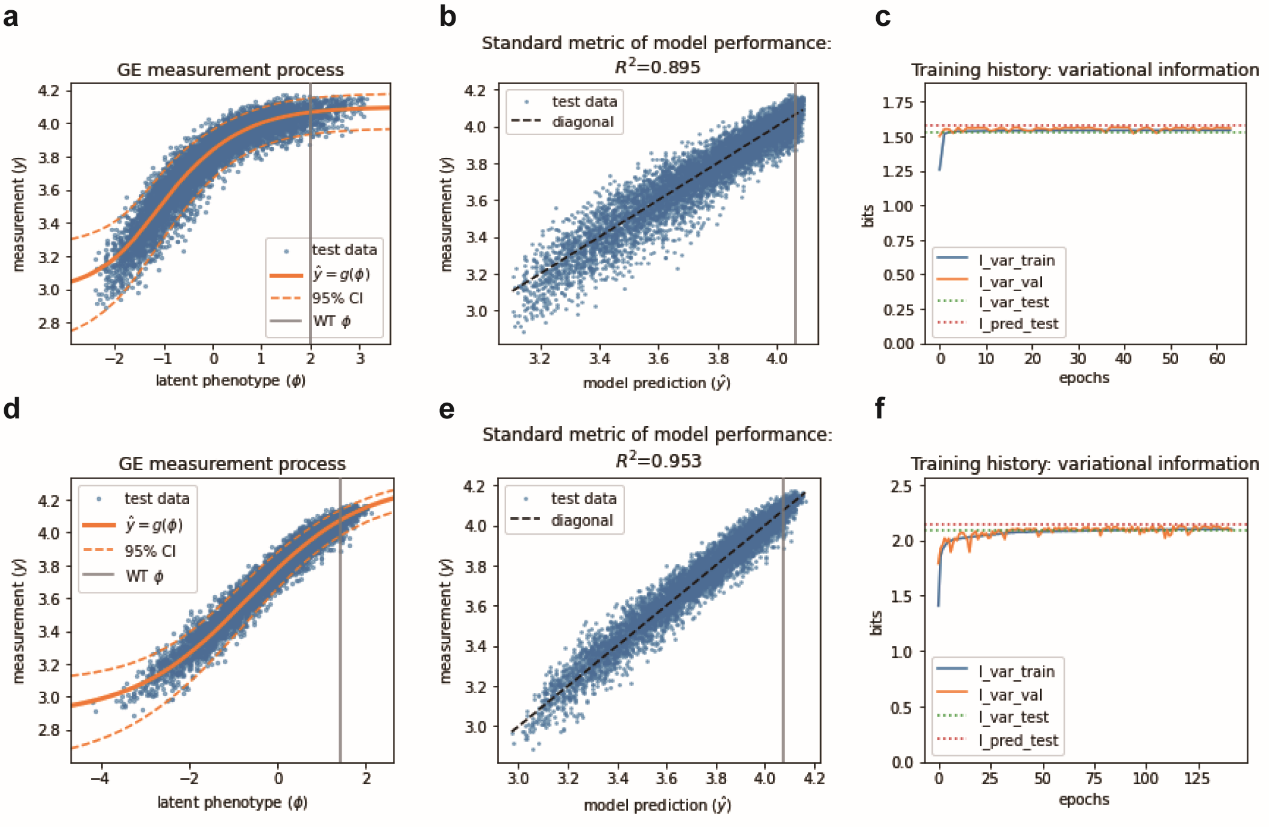
Figure 3 Modeling and phenotypic prediction of nonspecific epistasis of CreiLOV: (a-c) Based on G-P map additive model; (d-f) Based on G-P map black box model
As mentioned earlier, in order to obtain a better phenotype, it is often necessary to introduce multi-loci combinatorial mutagenesis into amino acid sequences, but the combinatorial explosion problem will pose great challenges for both rational design and experimental testing. In order to explore whether low-order mutant datasets can be used to predict high-order mutation combination effects, the author trained the MAVE-NN machine learning model using order 1/2/3/4/5 mutant data. It was found that the Pearson correlation coefficient between the model prediction and experimental results reached 0.84 when a dataset of order 3 or below mutants was used to predict mutagenic effects above order 6. It is worth noting that relatively accurate predictions were achieved using only a 10% subset of the order 1-3 mutant dataset (The Pearson correlation coefficient was 0.79) (Figure 4). Furthermore, the author used another machine learning model ECNet and other combinatorial mutagenesis datasets (CR9114 and avGFP) reported in the literature to explore the universality and limiting factors of using low-order mutation data to predict high-order mutation combination effects.

Figure 4 Prediction of phenotypes of high-order mutants of CreiLOV: (a) Prediction of phenotypes of order 6 and above mutants using order 1/2/3/4/5 and below low-order mutant datasets; (b) Prediction of phenotypes of order 6 and above mutants using data of order 3 and below mutants with different proportions
Finally, the author performed multiple rounds of FACS screening on single locus and combinatorial saturation mutagenesis libraries, and obtained multiple single locus and multi-loci mutants. Their in vivo fluorescence intensity and in vitro fluorescence quantum yield were significantly improved (The maximum fluorescence quantum yield reached 0.57), and their thermal stability was also improved at 60℃ (Figure 5), which has potential application value.
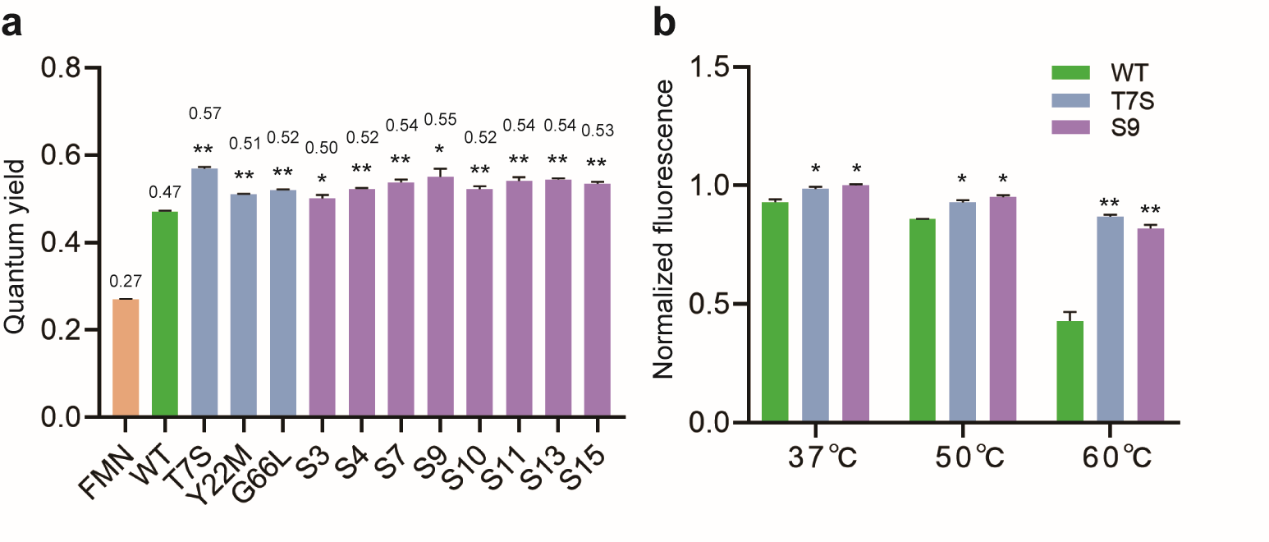
Figure 5 Characterization of dominant mutants of CreiLOV: (a) Fluorescence quantum yield; (b) Thermal stability
In summary, the paper conducted deep mutational scanning for single locus and multi-loci combinatorial saturation mutagenesis libraries, depicted the mutagenic effect and epistasis of amino acids of CreiLOV, and screened CreiLOV mutants with significantly improved performance. In addition, the author also demonstrated the feasibility of prediction of high-order mutant phenotypes based on a small amount of low-order mutant data by a machine learning model, which provided important reference and guidance for machine learning-aided protein engineering optimization design.
This achievement was supported by the National Key Research and Development Program (2020YFA090023 and 2021YFA0910800), the National Natural Science Foundation (32071428), the Guangdong Basic and Applied Basic Research Foundation (2021A1515110722), and Shenzhen Institute of Synthetic Biology. The author particularly thanks researcher Dai Lei of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, for his discussion on DMS data analysis, and Professor Zhang Chong of Tsinghua University for his help in FACS.