
On April 12, Beijing time, Ma Yingfei's research group and Dai Lei's research group of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published the latest research achievement in Cell Host & Microbe under the title of “Large-scale phage cultivation for commensal human gut bacteria”. This work established for the first time a large-scale phage culture group technology for main commensal intestinal bacteria, used phage culture to analyze the mechanism of long-term coexistence of intestinal bacteria and phages, and demonstrated the application potential of these phages in the regulation of intestinal flora.
Screenshot of the published paper
Link of the paper: https://doi.org/10.1016/j.chom.2023.03.013
Studies have shown that phages are the most diverse but still not accurately recognized component in the human microecosystem. The interaction between these phages and their host bacteria affects human health. The use of phage therapy to precisely target and regulate intestinal flora is currently a frontier field in the study of gut microbe. Given the complex relationship between phages and bacteria, the analysis of the structure, composition, and function of phages in the gut microflora is crucial for developing phage therapy targeting important intestinal pathogenic bacteria. In addition, metagenomic studies have showed that bacteria and phages in the gut can coexist stably for a long time. What is the mechanism by which they can coexist stably? In the above studies, how to obtain phage culture of intestinal bacteria is an urgent problem in the current research of human gut microbiome.
The research team developed a series of phage isolation and culture techniques for common commensal gut bacteria. In the 411 strains of commensal gut bacteria tested from healthy people, they successfully obtained 209 non-redundant phages that could infect 42 types of bacteria, respectively. These bacteria included 15 Bacteroides, 19 Firmicutes, 4 Actinomycetes, and 4 Proteobacteria. These 209 phages had genome sizes ranging from 12kb to 168kb, and over 80% of them belonged to taxonomic genera that have not yet been described, indicating their high novelty and diversity.
Interestingly, these cultivable phages contained a class of tailed phages with minimum genomes, which ranged in size from 10 to 20 kbp and formed an independent branch in classification. Their host bacteria are all Gram-positive bacteria, including bifidobacteria and Clostridia that are widespread in the human gut. According to the ICTV classification criteria for tailed phages, all these small-genome phages are classified into the Salasmaviridae viridae, belonging to two subfamilies and 83 genera, respectively. SEM photos showed that the Salasmaviridae phage was a short-tailed phage (Figure 1) with a head diameter of 30-50nm. Its genome had a similar organization structure and conserved orthologous proteins, such as DNA polymerase. In addition, it was found that the group represented by two strains of phages, namely PD491P1 and AS73P1, was more widely distributed in the human population than crAss-like phages (crAss-like phages are considered the most widely distributed phages in the human gut) (Figure 2). The host bacteria of these phages belong to the Bacteroidales, including Bacteroides, Alistipes, Odoribacter, Parabacteroide, Prevotella, Barnesiella and other main members of Bacteroides in the human gut. However, they are significantly different from the phages of other such isolated bacteria, forming an independent branch. According to the ICTV classification criteria for tailed phages, the research team classified these phages into a new family of phages, tentatively named Paboviridae.

Figure 1: Discovery and characterization of small genome phages in the human gut
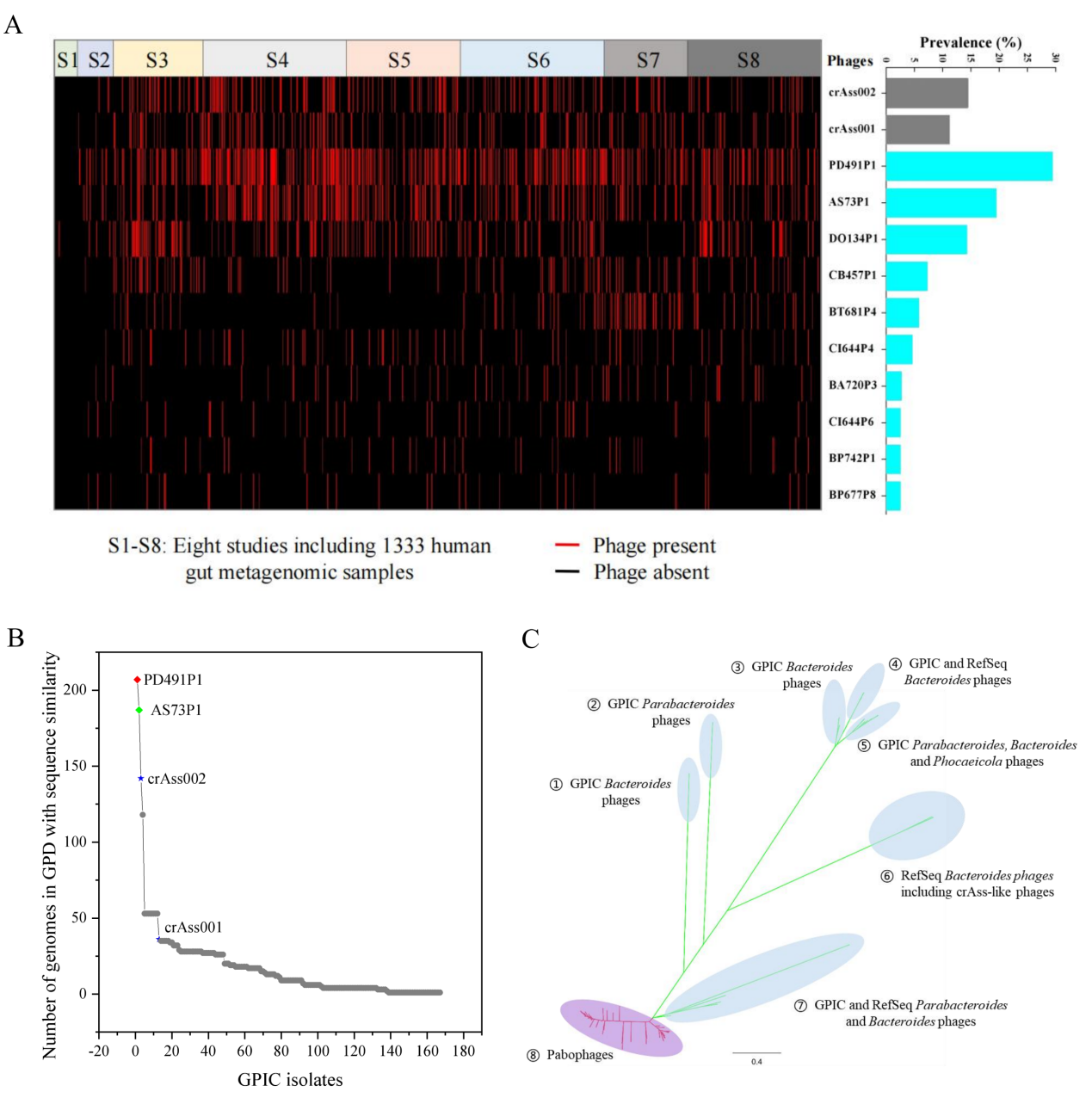
Figure 2: Discovery of a new phage family Paboviridae with high prevalence in the human gut
Previous bioinformatics analysis has shown that about one-third of human intestinal phages may have a wide host range. The research team selected phages of Bacteroides and Parabacteroides to test their host range. In thousands of cross-infection tests, no cross-species infection of phages on the host was observed, indicating that these phages had high host specificity (Figure 3A). Moreover, it was found that these bacteria from the same species also exhibited diverse phage sensitivity at the strain level, even though they came from fecal samples from the same person (Figure 3B), indicating that intestinal bacteria followed complex and diverse phage resistance mechanisms at the strain level. Therefore, the diverse phage resistance mechanisms of intestinal bacteria at the strain level was an important reason for the long-term stable coexistence of intestinal bacteria and phages.
Finally, to demonstrate the feasibility of using GPIC phages to regulate human intestinal flora, the team selected 8 bacteroides fragilis phages with different genome sequences and different host specificities to form a phage cocktail with a broad host range. In the in vitro culture system of intestinal flora in fecal samples from three donors, after phages were added for 24 hours, the abundance of bacteroides fragilis of all the samples significantly decreased, accompanied by drastic changes in the composition of the flora (Figure 3C and 3D). In longer continuous culture, phage cocktails could also reduce the abundance of bacteroides fragilis (Figures 3E and 3F), showing a persistent knockdown effect on the target species.

Figure 3: Host specificity and flora regulation of human intestinal phages
In summary, this study constructed a phage resource bank for main commensal bacteria in the gut, demonstrating the value of these phage cultures in regulating the intestinal flora. The study on the interaction between intestinal phages and bacteria to be conducted using this resource will help to deeply understand the diversity, function, and role of intestinal microbiome, and provide new ideas and methods for the prevention and treatment of related diseases.
Researcher Ma Yingfei and Research Dai Lei of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, are the co-corresponding authors of the paper, and assistant researcher Shen Juntao is the first author of the paper. This research work was conducted based on Shenzhen Major Technological Infrastructure for Synthetic Biology Research, and greatly supported by Li Bing's team of Tsinghua Shenzhen International Graduate School and Zhang Tong's team of the University of Hong Kong.
This work was sponsored by programs such as the National Key Research and Development Program (2019YFA0906700), the Priority Research Program of the Chinese Academy of Sciences (XDB29050501), the National Natural Science Foundation of China (31971513, 32061143023), the Guangdong Basic and Applied Basic Research Foundation (2020A1515110184), and Shenzhen Institute of Synthetic Biologyn.