
On March 18, Beijing time, Lou Chunbo's research group of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Ouyang Qi/Qian Long's team of the Quantitative Biology Center of the School of Physics, Peking University, jointly published on Nature Communications an artificial genetic circuit that precisely controls the expression dose of multiple genes in mammalian cells under the title of “Precise programming of multigene expression stoichiometry in mammalian cells by a modular and programmable transcriptional system”. In this work, by constructing an artificial orthogonal transcriptional system in mammals, precise fine-tuning of the transcriptional activity of single and multiple promoters was realized, and a quantitative thermodynamic model of multigene expression with predictive ability was constructed. This quantitative model was applied to the optimization design of the composition and yield of virus-like particles (VLP) for influenza A virus (H1N1). Qin Chenrui, Xiang Yanhui, and Liu Jie are co-first authors, and Lou Chunbo of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Qian Long of Peking University are the co-corresponding authors of the paper.
Screenshot of the published paper
Link of the paper: https://www.nature.com/articles/s41467-023-37244-y
In mammalian cells, precise regulation of genetic circuits is crucial for physiological functions such as environmental adaptation, homeostasis, development and differentiation of cells. Overexpression or underexpression of key genes can lead to major diseases such as cancer. In addition, the expression dose of multiple cell fate determinants is also a key factor in reshaping cell fate differentiation and development. However, in mammalian cells, gene expression is affected by a number of complex factors, such as gene order, genomic location, epigenetic modifications, and host cell type, making it very difficult to precisely control of the expression dose of genes. For example, the transcriptional activity of the commonly used promoter EF1α is significantly influenced by cell type in seven cell lines such as HEK293T; while the expression activity of CMV strong promoter in motor neuron cells may be very high at the beginning and then gradually weakened. Therefore, modular, cell-type independent and programmable gene expression systems have become an important bottleneck in mammalian cell biology and synthetic biology research.
This paper proposed a design strategy for developing a modular, host-independent orthogonal transcriptional system. The orthogonal transcriptional system consisted of an orthogonal promoter library and monomeric RNA polymerase (RNAP). By fusing RNA-capping enzymes with the monomeric RNAP, the system ensured that prokaryotic monomeric RNAPs performed the necessary steps of eukaryotic protein expression in mammalian cells, such as gene transcription, posttranscriptional modification, nuclear export, and translation, in a “cross-domain” manner (Figure 1).

Figure 1. Construction of the orthogonal promoter library and the cross-species universal transcriptional system and experimental verification of the quantitative model
In this paper, the competition effect of expression activity of different genes was discovered and a quantitative thermodynamic model was established. Aimed at the competition issue between two genes, the research team designed two reporter genes in a mammalian cell line (Figure 2). In this dual reporter gene system, each reporter gene was controlled by one of the seven representative promoters in the orthogonal promoter library. There were a total of 49 different combinations. The experimental results showed that the strong promoter of one gene significantly reduced the expression of the other gene. This result proved that two reporter genes were competing for limited resources ([RNAP]free). Therefore, in this paper, a new thermodynamic equation using [RNAP]free to replace [RNAP]tot was proposed (Figure 2e).

Figure 2. Competition effects of the activity of multiple promoters caused by the sharing of key factors in mammalian cells and the related quantitative thermodynamic model
In mammalian cells, the order of multiple genes also significantly affected gene expression. To investigate the impact of gene order on gene expression, the research team designed six possible orders of three reporter genes (1-2-3, 1-3-2, 2-1-3, 2-3-1, 3-1-2, 3-2-1) and calculated the variance of the expression of the reporter genes in all six combinations (Figure 3). The authors found that the coefficient of variance (CV) of the expression of the same promoter was very small. This result suggested that the promoter activity was not significantly affected by its local genetic environment, and similar behavior was maintained in both CHO and HEK293T cell lines. In addition, the research team randomly selected promoters for three different reporter genes in 50 promoter libraries (with activity changes by about 100 times). Through a modified thermodynamic model, the paper predicted the activity of three reporter genes. According to the experimental results, it was found that the protein expression of all three genes in mammalian cells can be predicted by the model (R2=0.81~0.88).
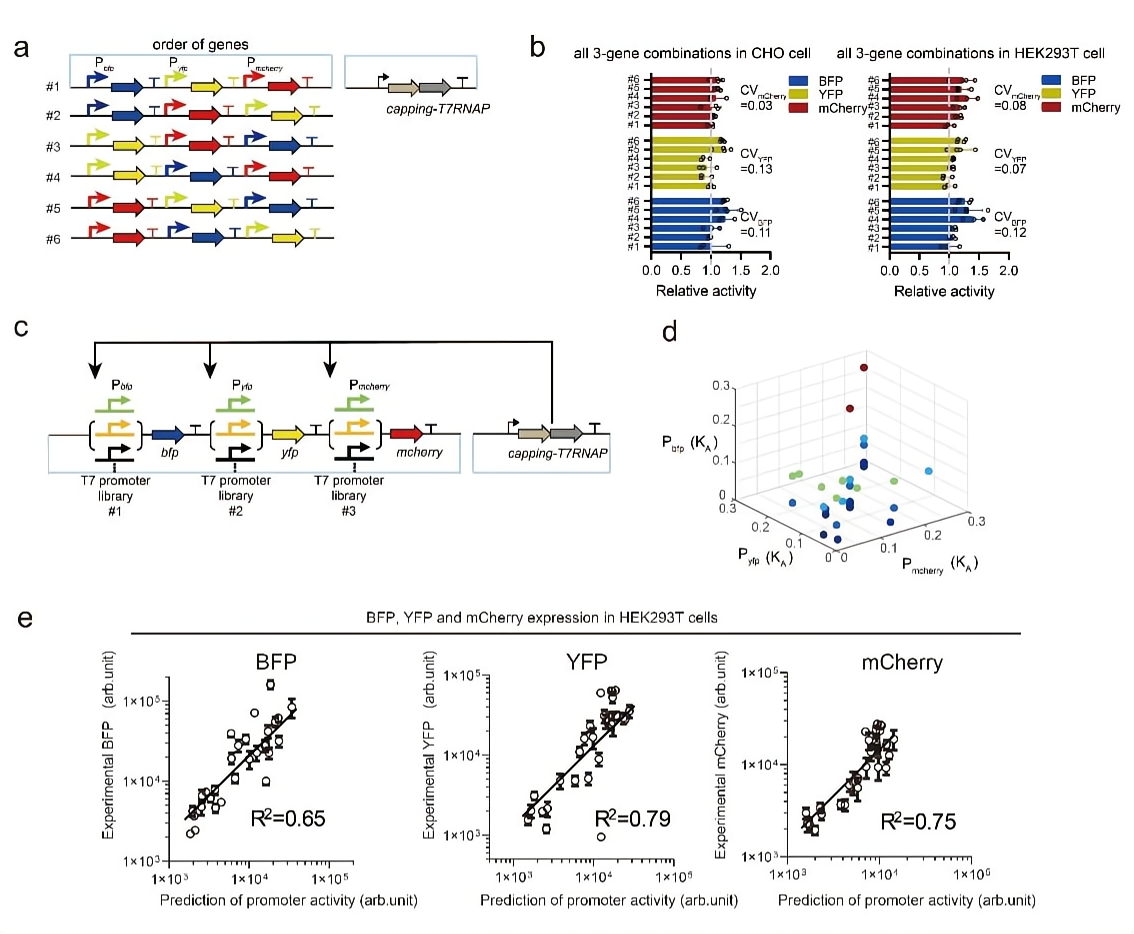
Figure 3. The small impact of gene order on the orthogonal transcriptional system and the predictability of the related quantitative thermodynamic model
To demonstrate the benefits of the orthogonal transcriptional system, the research team optimized the gene expression dose of multiple subunits of virus-like particles using the quantitative thermodynamic model. Viral-like particles (VLP) are non-pathogenic nanoparticles composed of multiple proteins that cannot replicate. They have a wide range of potential applications in biomedical fields such as vaccines and drug delivery. The expression of multiple protein subunits is crucial for the assembly efficiency and immunogenicity of VLP. Therefore, the team investigated the programmability of expression and its impact on VLP yield. First, the authors validated that under appropriate expression dose conditions, the three key subunits of influenza A virus (hemagglutinin (HA), neuraminidase (NA), and matrix protein 1 (M1)) can form stable VLP. Then, they constructed two fusion genes (egfp-M1 and NA-mCherry) to quantitatively characterize the yield and integrity of VLP. Subsequently, they predicted the VLP yield for all preset promoter parameters and gene-specific parameters (a total of 157,464 groups) by the revised quantitative thermodynamic model. Through virtual screening and theoretical analysis, the research team found that high expression of the HA gene was harmful to the production of VLP, while the three genes with appropriate expression can produce more VLP particles. Finally, the team selected more than ten high-yield combinations for experimental verification, and found that all combinations produced more VLP particles, and the experimental results were consistent with the predicted values. The higher the predicted value, the better the experimental results.

Figure 4. Predictable optimization and experimental verification of the expression dose of the three subunit genes of virus-like particles (VLP) of influenza A virus
In summary, in this study, a modular and programmable orthogonal transcriptional regulatory system was developed. In addition, the study also established a quantitative thermodynamic theoretical model based on intracellular resource competition and binding energy, which realized precise design and prediction of the expression dose of multiple genes in mammalian cells. By utilizing this orthogonal transcriptional system, the research team also successfully optimized the virus-like particles (VLP) of influenza A virus, providing potential for the development and production of new and efficient influenza A vaccines.
This work was sponsored by the National Key Research and Development Program, the National Natural Science Foundation of China, the Priority Program of the Chinese Academy of Sciences, the Youth Cross Team Program, and Shenzhen Institute of Synthetic Biology.