
One of the core visions of synthetic biology is the artificial synthesis of single-celled life. To achieve this goal, researchers need to precisely control the timing of cell division to ensure that the size of each generation of cells remains within a certain range. If the cells do not divide for too long, the cell volume will get larger and larger, the error rate of DNA replication and isolation processes will increase, and cell metabolism will also be affected. Conversely, if the cells divide prematurely, it may lead to small cell volume, insufficient cytoplasm, or even incomplete DNA replication after division, resulting in the cessation of life activities.
Bacteria are rapidly growing microorganisms whose cell volume can double within 20 minutes. To achieve rapid growth, bacteria do not set checkpoints to clearly distinguish processes such as cell growth and chromosome replication, as eukaryotes do. In this case, how do bacteria determine when to divide? Although humans have viewed bacteria for hundreds of years, they have not fully understood the mystery of the mechanism yet.
On March 14, Beijing time, Fu Xiongfei, a researcher, and Bai Yang, an associate researcher, of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, jointly with Dr. Luo Liang of Huazhong Agricultural University published a research article in Physical Review Research, pointing out that random fluctuations at the molecular level are important factors affecting the regulation of bacterial cell division.
Screenshot of the published paper
Link of the paper: https://link.aps.org/doi/10.1103/PhysRevResearch.5.013173
At present, there are three possible regulatory mechanisms that can describe bacterial cell division. The first is the volume determining mechanism, which assumes that the cell monitors its own volume and performs cell division when it reaches a specific division threshold. The second is the increment determining mechanism, in which the cell records the volume increase since the last division and performs division when the increment reaches a specific division threshold. The last is the time determining mechanism, in which the cell records the time elapsed since the last division and performs division when the time elapsed reaches a specific division threshold. However, scientists have not been able to figure out exactly which determining mechanism is used for bacteria cell division, and what is the molecular basis behind it.
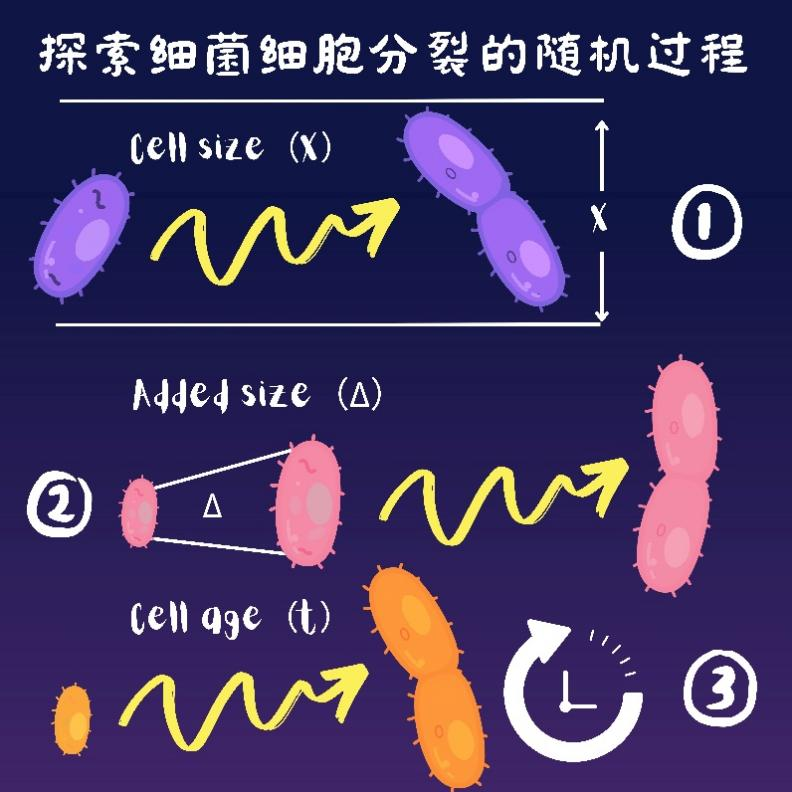
Figure: Three determining mechanisms of bacterial cell division
The importance of cell division timing for cell growth and genetic stability is evident. Scientists have long been exploring the determining mechanism of cell division timing. In recent years, scientists have realized long-term photographic observations of individual bacterial cells, recording the process of birth - growth - division of multiple generations of cells. Through these observations, they found an interesting phenomenon: Although the volume of bacteria varies, on average, the volume increase between two divisions does not vary with the volume of the bacteria at birth. This discovery seems to indicate that the bacterial cell determines the timing of division with volume increment as the regulatory target. This discovery has triggered a new wave of research on the regulatory mechanisms of bacterial cell division, which greatly deepened our understanding of bacterial cell cycle. However, new research has raised doubts about the mechanism by which volume increment determines the timing of cell division. Scientists have yet to identify the determining mechanism of the timing of bacterial cell division.
In the newly published paper, Fu Xiongfei's research group pointed out that people may have misunderstood the true meaning behind the experimental observation that “the volume increment of bacteria does not vary with its volume at birth”. This is because the ubiquitous stochastic process in the cell also acts on the cell division threshold, so that all three different cell division determining mechanisms can show an experimental phenomenon of “volume increment does not vary with the volume of bacteria at its birth”. By constructing a theoretical model that includes randomness, this study fully analyzed the impact of stochastic process on cell division behavior. The study has found that with the different characteristics of the stochastic process of the division threshold (autocorrelation coefficient), all three determining mechanisms of cell division timing can show experimental phenomena that seem to be volume determination, increment determination, or time determination. Therefore, researchers need to further measure the autocorrelation coefficient of the division threshold itself in order to more accurately understand the regulatory mechanism of bacterial cell division.
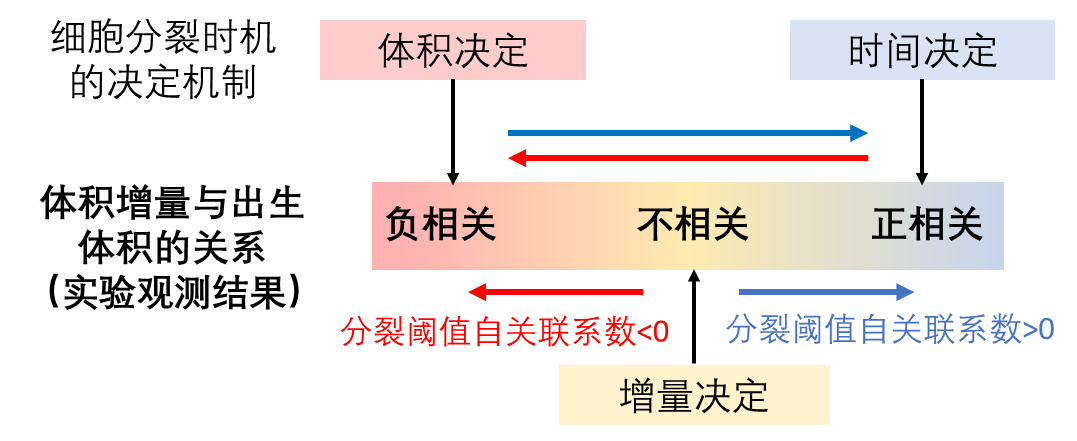
Figure: The stochastic process of bacteria division threshold affects the characteristics of cell division behavior, so that the same division determining mechanism exhibits diverse experimental observations
This discovery greatly improves the flexibility of rational design of artificial cells. Researchers can control the division behavior of bacterial cells through the ingenious design of the stochastic process characteristics of the division threshold under the circumstance that the determining mechanism of cell division is difficult to modify. This work was supported by the National Key Research and Development Program of the Ministry of Science and Technology, the Priority Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China, and Shenzhen Institute of Synthetic Biology.
PI introduction:
Fu Xiongfei, researcher, doctoral supervisor, deputy director of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, deputy director of the Key Laboratory of Quantitative Engineering Biology, Chinese Academy of Sciences, and vice president of Shenzhen Institute of Synthetic Biology. The main research direction of the research group: By combining quantitative theory and synthetic reconstruction, conduct research on issues such as the interaction between intracellular gene circuits and chassis, and the formation principle of unicellular and multicellular ordered spatial structure.
The research group is now recruiting postdoctoral fellows, research assistants, technicians, visiting students, etc. with relevant research backgrounds in mathematics, physics, chemistry, biology, fluid mechanics, complex systems, etc. If you are interested, please contact yang.bai@siat.ac.cn. The subject line of the resume and email should be “Position applied - School Name - Major - Name”.
Laboratory homepage:
http://isynbio.siat.ac.cn/fulab